smilite is a Python module to download and analyze SMILES strings (Simplified Molecular-Input Line-entry System) of chemical compounds from ZINC (a free database of commercially-available compounds for virtual screening, http://zinc.docking.org).
Now supports both Python 3.x and Python 2.x.
• Installation
• Simple command line online query scripts
- lookup_zincid.py
- lookup_smile_str.py
• CSV file command line scripts
- gen_zincid_smile_csv.py (downloading SMILES)
- comp_smile_strings.py (checking for duplicates within 1 file)
- comp_2_smile_files.py (checking for duplicates across 2 files)
• SQLite file command line scripts
- lookup_single_id.py
- lookup_smile.py
- add_to_sqlite.py
- sqlite_to_csv.py
• Changelog
You can use the following command to install smilite:
pip install smilite
or
easy_install smilite
Alternatively, you can download the package manually from the Python Package Index https://pypi.python.org/pypi/smilite, unzip it, navigate into the package, and use the command:
python3 setup.py install
If you downloaded the smilite package from https://pypi.python.org/pypi/smilite or https://github.com/rasbt/smilite, you can use the command line scripts I provide in the scripts/cmd_line_online_query_scripts dir.
Retrieves the SMILES string and simplified SMILES string for a given ZINC ID
from the online Zinc. It uses ZINC12 as the default backend, and via an additional commandline argument zinc15, the ZINC15 database will be used instead.
Usage:
[shell]>> python3 lookup_zincid.py ZINC_ID [zinc12/zinc15]
Example (retrieve data from ZINC):
[shell]>> python3 lookup_zincid.py ZINC01234567 zinc15
Output example:
ZINC01234567 C[C@H]1CCCC[NH+]1CC#CC(c2ccccc2)(c3ccccc3)O CC1CCCCN1CCCC(C2CCCCC2)(C3CCCCC3)O
Where
- 1st row: ZINC ID
- 2nd row: SMILES string
- 3rd row: simplified SMILES string
Retrieves the corresponding ZINC_IDs for a given SMILES string
from the online ZINC database.
Usage:
[shell]>> python3 lookup_smile_str.py SMILE_str
Example (retrieve data from ZINC):
[shell]>> python3 lookup_smile_str.py "C[C@H]1CCCC[NH+]1CC#CC(c2ccccc2)(c3ccccc3)O"
Output example:
ZINC01234567 ZINC01234568 ZINC01242053 ZINC01242055
If you downloaded the smilite package from https://pypi.python.org/pypi/smilite or https://github.com/rasbt/smilite, you can use the command line scripts I provide in the scripts/csv_scripts dir.
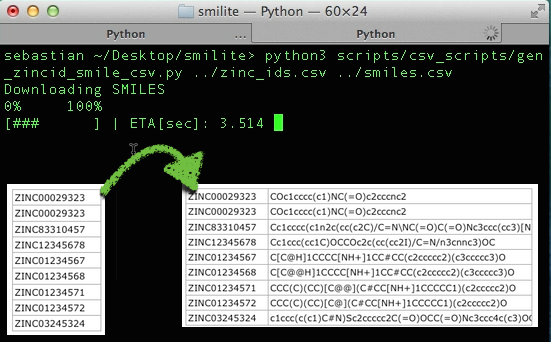
Generates a ZINC_ID,SMILE_STR csv file from a input file of ZINC IDs. The input file should consist of 1 columns with 1 ZINC ID per row. ZINC12 is used as the default backend, and via an additional commandline argument zinc15, the ZINC15 database can be used instead.
Usage:
[shell]>> python3 gen_zincid_smile_csv.py in.csv out.csv [zinc12/zinc15]
Example:
[shell]>> python3 gen_zincid_smile_csv.py ../examples/zinc_ids.csv ../examples/zid_smiles.csv zinc15
Screen Output:
Downloading SMILES 0% 100% [########## ] | ETA[sec]: 106.525
Input example file format:
zinc_ids.csv
Output example file format:
zid_smiles.csv
Compares SMILES strings within a 2 column CSV file (ZINC_ID,SMILE_string) to identify duplicates. Generates a new CSV file with ZINC IDs of identified duplicates listed in a 3rd-nth column(s).
Usage:
[shell]>> python3 comp_smile_strings.py in.csv out.csv [simplify]
Example 1:
[shell]>> python3 comp_smile_strings.py ../examples/zinc_smiles.csv ../examples/comp_smiles.csv
Input example file format:
zid_smiles.csv
Output example file format 1:
comp_smiles.csv
Where
- 1st column: ZINC ID
- 2nd column: SMILES string
- 3rd column: number of duplicates
- 4th-nth column: ZINC IDs of duplicates
Example 2:
[shell]>> python3 comp_smile_strings.py ../examples/zid_smiles.csv ../examples/comp_simple_smiles.csv simplify
Output example file format 2:
comp_simple_smiles.csv
Compares SMILES strings between 2 input CSV files, where each file consists of rows with 2 columns ZINC_ID,SMILE_string to identify duplicate SMILES string across both files.
Generates a new CSV file with ZINC IDs of identified duplicates listed in a 3rd-nth column(s).
Usage:
[shell]>> python3 comp_2_smile_files.py in1.csv in2.csv out.csv [simplify]
Example:
[shell]>> python3 comp_2_smile_files.py ../examples/zid_smiles2.csv ../examples/zid_smiles3.csv ../examples/comp_2_files.csv
Input example file 1:
zid_smiles2.csv
Input example file 2:
zid_smiles3.csv
Output example file format:
comp_2_files.csv
Where:
- 1st column: name of the origin file
- 2nd column: ZINC ID
- 3rd column: SMILES string
- 4th-nth column: ZINC IDs of duplicates
If you downloaded the smilite package from https://pypi.python.org/pypi/smilite or https://github.com/rasbt/smilite, you can use the command line scripts I provide in the scripts/sqlite_scripts dir.
Retrieves the SMILES string and simplified SMILES string for a given ZINC ID
from a previously built smilite SQLite database or from the online ZINC database.
Usage:
[shell]>> python3 lookup_single_id.py ZINC_ID [sqlite_file]
Example1 (retrieve data from a smilite SQLite database):
[shell]>> python3 lookup_single_id.py ZINC01234567 ~/Desktop/smilite_db.sqlite
Example2 (retrieve data from the ZINC online database):
[shell]>> python3 lookup_single_id.py ZINC01234567
Output example:
ZINC01234567 C[C@H]1CCCC[NH+]1CC#CC(c2ccccc2)(c3ccccc3)O CC1CCCCN1CCCC(C2CCCCC2)(C3CCCCC3)O
Where
- 1st row: ZINC ID
- 2nd row: SMILES string
- 3rd row: simplified SMILES string
Retrieves the ZINC ID(s) for a given SMILES string or simplified SMILES string from a previously built smilite SQLite database.
Usage:
[shell]>> python3 lookup_smile.py sqlite_file SMILE_STRING [simplify]
Example1 (search for SMILES string):
[shell]>> python3 lookup_smile.py ~/Desktop/smilite.sqlite "C[C@H]1CCCC[NH+]1CC#CC(c2ccccc2)(c3ccccc3)O"
Example2 (search for simplified SMILES string):
[shell]>> python3 lookup_smile.py ~/Desktop/smilite.sqlite "CC1CCCCN1CCCC(C2CCCCC2)(C3CCCCC3)O" simple
Output example:
ZINC01234567 C[C@H]1CCCC[NH+]1CC#CC(c2ccccc2)(c3ccccc3)O CC1CCCCN1CCCC(C2CCCCC2)(C3CCCCC3)O
Where
- 1st row: ZINC ID
- 2nd row: SMILES string
- 3rd row: simplified SMILES string
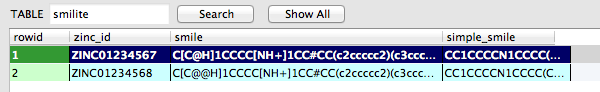
Reads ZINC IDs from a CSV file and looks up SMILES strings and simplified SMILES strings from the ZINC online database. Writes those SMILES strings to a smilite SQLite database. A new database will be created if it doesn't exist, yet.
Usage:
[shell]>> python3 add_to_sqlite.py sqlite_file csv_file
Example:
[shell]>> python3 add_to_sqlite.py ~/Desktop/smilite.sqlite ~/Desktop/zinc_ids.csv
Input CSV file example format:
ZINC01234567 ZINC01234568 ...
An example of the smilite SQLite database contents after successful insertion is shown in the image below.
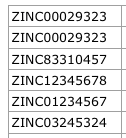
Writes contents of an SQLite smilite database to a CSV file.
Usage:
[shell]>> python3 sqlite_to_csv.py sqlite_file csv_file
Example:
[shell]>> python3 sqlite_to_csv.py ~/Desktop/smilite.sqlite ~/Desktop/zinc_smiles.csv
Input CSV file example format:
ZINC_ID,SMILE,SIMPLE_SMILE ZINC01234568,C[C@@H]1CCCC[NH+]1CC#CC(c2ccccc2)(c3ccccc3)O,CC1CCCCN1CCCC(C2CCCCC2)(C3CCCCC3)O ZINC01234567,C[C@H]1CCCC[NH+]1CC#CC(c2ccccc2)(c3ccccc3)O,CC1CCCCN1CCCC(C2CCCCC2)(C3CCCCC3)O
An example of the CSV file contents opened in an spreadsheet program is shown in the image below.
VERSION 2.3.1 (07/25/2020)
- Fix bug to allow
zinc15option in gen_zincid_smile_csv.py script
VERSION 2.3.0 (06/10/2020)
- Fixes ZINC URL in
lookup_smile_str.py - Adds an optional command line parameter (with arguments
zinc15orzinc12) forlookup_smile_str.py
VERSION 2.2.0
- Provides an optional command line argument (zinc15) to use ZINC15 as a backend for downloading SMILES
VERSION 2.1.0
- Functions and scripts to fetch ZINC IDs corresponding to a SMILES string query
VERSION 2.0.1
- Progress bar for add_to_sqlite.py
VERSION 2.0.0
- added SQLite features
VERSION 1.3.0
- added script and module function to compare SMILES strings across 2 files.
VERSION 1.2.0
- added Python 2.x support
VERSION 1.1.1
- PyPrind dependency fix
VERSION 1.1.0
- added a progress bar (PyPrind) to
generate_zincid_smile_csv()function